Two sample permutation test of respiratory phase angles#
This notebook runs permutation tests to compare two samples of respiratory phase angles. This is useful when you want to compare the phase angles of two different conditions or groups.
[1]:
import pickle
import numpy as np
from pathlib import Path
import matplotlib.pyplot as plt
from pyriodic.viz import CircPlot
from pyriodic.permutation import permutation_test_between_samples
Single-level analysis#
[2]:
data_path = Path("../../data/respiration/intermediate")
subj_id = "0019"
file_path = data_path / f"participant_{subj_id}_preproc.pkl"
with open(file_path, 'rb') as f:
data = pickle.load(f)
circ = data["circ"]
PA = data["phase_angles"]
Choosing events of interest and plotting the two samples#
[3]:
targets_weak = circ["weak/target"]
[4]:
correct_targets = targets_weak["correct"]
incorrect_targets = targets_weak["incorrect"]
fig, axes = plt.subplots(1, 2, dpi = 300, figsize = (10, 6), subplot_kw={"projection": "polar"}, sharey=True)
correct_plot = CircPlot(
correct_targets,
title = "Correct targets",
group_by_labels=False,
ax=axes[0]
)
correct_plot.add_points(s=3)
correct_plot.add_density()
correct_plot.add_histogram(PA)
incorrect_plot = CircPlot(
incorrect_targets,
title = "Incorrect targets",
group_by_labels=False,
ax=axes[1]
)
incorrect_plot.add_points(s=3)
incorrect_plot.add_density()
incorrect_plot.add_histogram(PA)
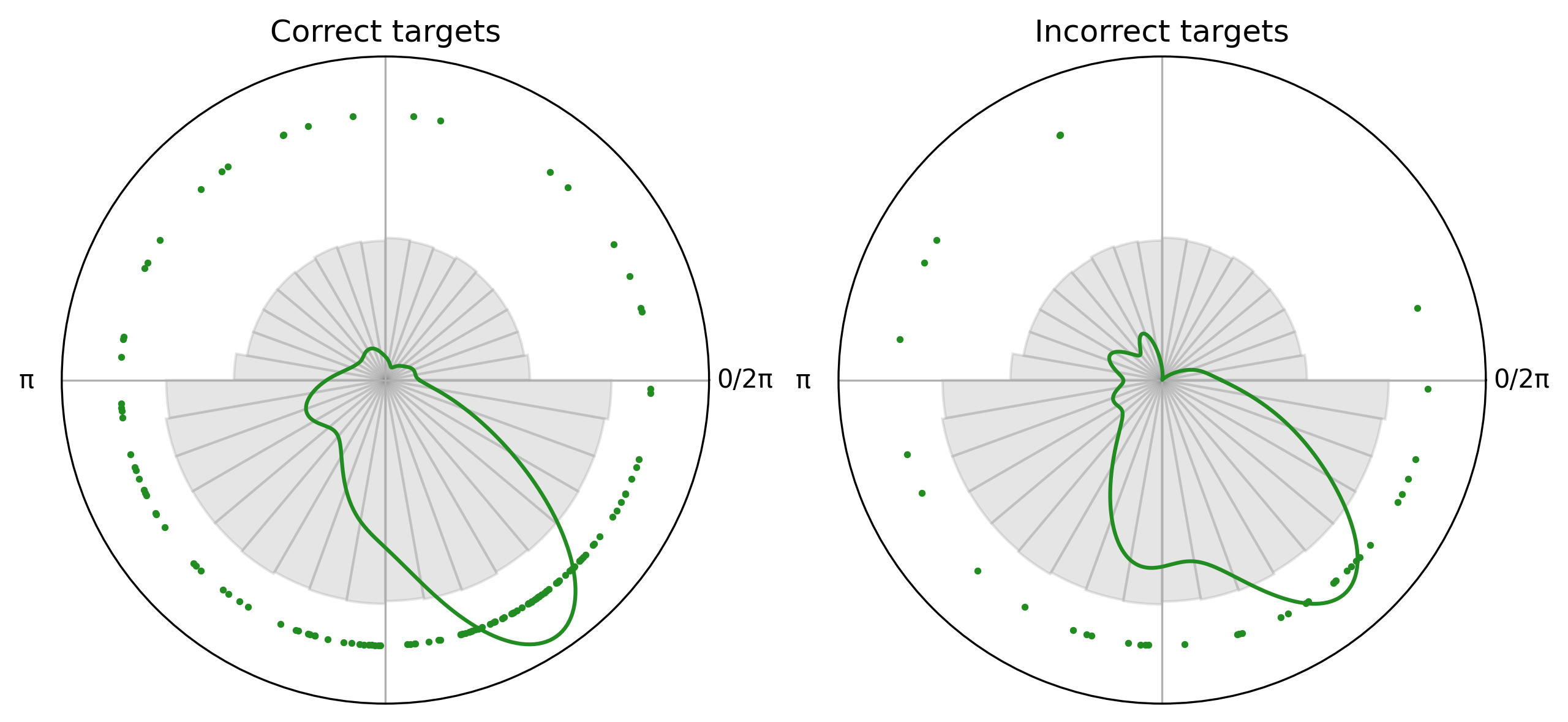
[5]:
# Run permutation test
obs_stat, pval = permutation_test_between_samples(
correct_targets.data,
incorrect_targets.data,
n_permutations=10000
)
Observed statistic = 0.040, p = 0.8397
[6]:
circs = {}
for file_path in data_path.iterdir():
subj_id = file_path.name.split("_")[1]
with open(file_path, 'rb') as f:
data = pickle.load(f)
circs[subj_id] = data["circ"]
n_permutations = 1000
group_observed_stats = []
group_null_stats = []
group_pvals = []
for subj_id in circs:
pa = circs[subj_id]
weak_targets = circs[subj_id]["weak/target"]
correct_targets_tmp = weak_targets["correct"]
incorrect_targets_tmp = weak_targets["irregular"]
# Run subject-level permutation test
obs, pval, null_distribution = permutation_test_between_samples(
incorrect_targets_tmp.data,
correct_targets_tmp.data,
n_permutations=n_permutations,
verbose=False,
return_null_distribution=True,
)
group_observed_stats.append(obs)
group_null_stats.append(null_distribution)
group_pvals.append(pval)
# Group-level observed statistic: average of subject-level observed stats
# On average, do subjects show evidence of difference in phase for irregular and regular targets?
group_obs = np.mean(group_observed_stats)
# Group-level null: average of null stats across subjects, per null_stat
# Reference distribution of what kind of group-level effect you’d expect if there was no difference between irregular and regular targets
group_null = [
np.mean([nulls[i] for nulls in group_null_stats])
for i in range(n_permutations)
]
# Group-level p-value
p_val = (np.sum(np.array(group_null) >= group_obs) + 1) / (len(group_null) + 1)
print(f"Group-level statistic = {group_obs:.3f}, p = {p_val:.4f}")
Group-level statistic = 0.024, p = 1.0000
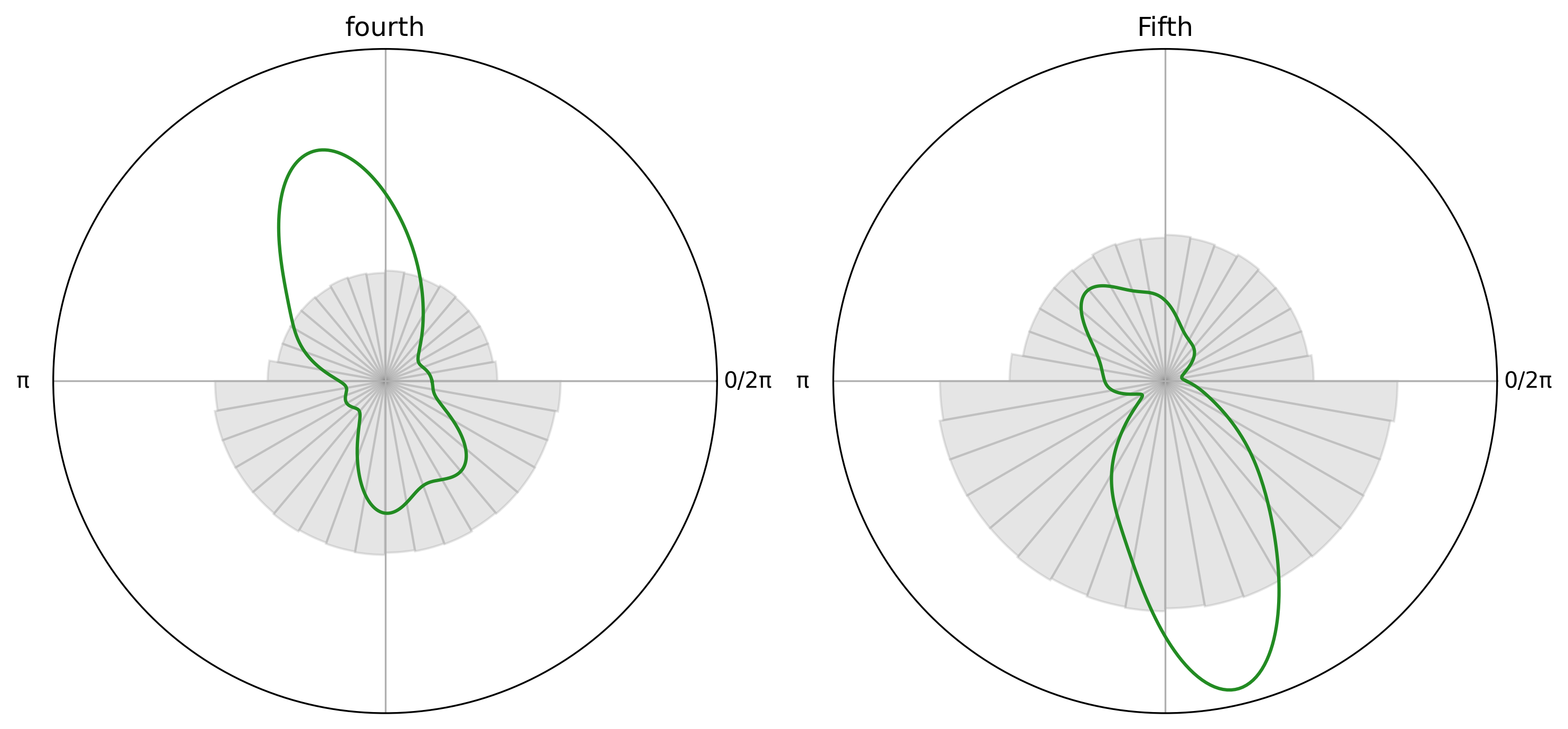
Another example where there is a difference between the two samples#
[ ]:
fourth_salient = circ["salient/regular/fourth"]
fifth_salient = circ["salient/regular/fifth"]
fig, axes = plt.subplots(1, 2, dpi = 300, figsize = (10, 6), subplot_kw={"projection": "polar"}, sharey=True, sharex=True)
fourth_plot = CircPlot(
fourth_salient,
title="fourth",
group_by_labels=False,
ax=axes[0]
)
fourth_plot.add_density()
fourth_plot.add_histogram(PA)
fifth_plot = CircPlot(
fifth_salient,
title="Fifth",
group_by_labels=False,
ax=axes[1]
)
fifth_plot.add_density()
fifth_plot.add_histogram(PA)
plt.tight_layout()
# Run permutation test
obs_stat, pval = permutation_test_between_samples(
fourth_salient.data,
fifth_salient.data,
n_permutations=10000,
)
Observed statistic = 0.748, p = 0.0001